Abstract
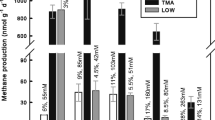
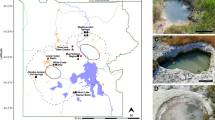
Culture-independent and enrichment techniques, with an emphasis on members of the Archaea, were used to determine the composition and structure of microbial communities inhabiting microbial mats in the source pools of two geothermal springs near the towns of Arzakan and Jermuk in Armenia. Amplification of small-subunit rRNA genes using “universal” primers followed by pyrosequencing (pyrotags) revealed highly diverse microbial communities in both springs, with >99 % of pyrosequences corresponding to members of the domain Bacteria. The spring in Arzakan was colonized by a photosynthetic mat dominated by Cyanobacteria, in addition to Proteobacteria, Bacteroidetes, Chloroflexi, Spirochaeta and a diversity of other Bacteria. The spring in Jermuk was colonized by phylotypes related to sulfur, iron, and hydrogen chemolithotrophs in the Betaproteobacteria and Epsilonproteobacteria, along with a diversity of other Bacteria. Analysis of near full-length small subunit rRNA genes amplified using Archaea-specific primers showed that both springs are inhabited by a diversity of methanogens, including Methanomicrobiales and Methanosarcinales and relatives of Methanomassiliicoccus luminyensis, close relatives of the ammonia-oxidizing archaeon (AOA) “Candidatus Nitrososphaera gargensis”, and the yet-uncultivated Miscellaneous Crenarchaeotal Group and Deep Hydrothermal Vent Crenarchaeota group 1. Methanogenic enrichments confirmed the predicted physiological diversity, revealing methylotrophic, acetoclastic, and hydrogenotrophic methanogenesis at 45 and 55 °C, but not 65 °C. This is one of only a few studies combining cultivation-independent and -dependent approaches to study archaea in moderate-temperature (37–73 °C) terrestrial geothermal environments and suggests important roles for methanogenic archaea and AOA in the carbon and nitrogen biogeochemical cycles in these environments.



Similar content being viewed by others
References
Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay JÉ, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedrós-Alió C, Bertilsson S (2012) Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci USA 109:17989–17994
Badalyan M (2000) Geothermal features of Armenia: a country update. In: Proceedings World Geothermal Congress (2000), pp. 71–75
Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, Fredricks HF, Elvert M, Kelly TJ, Schrag DP, Sogin ML, Brenchley JE, Teske A, House CH, Hinrichs KU (2006) Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA 103:3846–3851
Brock TD (1967) Micro-organisms adapted to high temperatures. Nature 214:882–885
Chapelle FH, O’Neill K, Bradley PM, Methé BA, Ciufo SA, Knobel LL, Lovley DR (2002) A hydrogen-based subsurface microbial community dominated by methanogens. Nature 415:312–315
Cole JK, Peacock JP, Dodsworth JA, Williams AJ, Thompson DB, Dong H, Wu G, Hedlund BP (2013) Sediment microbial communities in great boiling spring are controlled by temperature and distinct from water communities. ISME J 7:718–729
Costa KC, Navarro JB, Shock EL, Zhang CL, Soukup D, Hedlund BP (2009) Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13:447–459
de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA (2008) Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol 10:810–818
Dodsworth JA, Hungate BA, Hedlund BP (2011) Ammonia oxidation, denitrification and dissimilatory nitrate reduction to ammonium in two US Great Basin hot springs with abundant ammonia-oxidizing archaea. Environ Microbiol 13:2371–2386
Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M (2012) Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62:1902–1907
Eder W, Ludwig W, Huber R (1999) Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of kebrit deep, red sea. Arch Microbiol 172:213–218
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Edwards TA, Calica NA, Huang DA, Manoharan N, Hou W, Huang L, Panosyan H, Dong H, Hedlund BP (2013) Wide geographic range of thermophilic Nitrospira species. FEMS Microbiol Ecol. doi:10.1111/1574-6941.12117
Felsenstein J (1989) PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164–166
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hatzenpichler R (2012) Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol 78:7501–7510
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA 105:2134–2139
Hou W, Wang S, Dong H, Jiang H, Briggs BR, Peacock JP, Huang Q, Huang L, Wu G, Zhi X, Li W, Dodsworth JA, Hedlund BP, Zhang C, Hartnett HE, Dijkstra P, Hungate BA (2013) A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PLoS One 8:e53350
Huang Z, Hedlund BP, Wiegel J, Zhou J, Zhang CL (2007) Molecular phylogeny of uncultivated Crenarchaeota in great basin hot springs of moderately elevated temperature. Geomicrobiol J 24:535–542
Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R (2007) Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460–465
Jiang B, Parshina SN, van Doesburg W, Lomans BP, Stams AJ (2005) Methanomethylovorans thermophila sp. nov., a thermophilic, methylotrophic methanogen from an anaerobic reactor fed with methanol. Int J Syst Evol Microbiol 55:2465–2470
Kämpfer P, Schulze R, Jäckel U, Malik KA, Amann R, Spring S (2005) Hydrogenophaga defluvii sp. nov. and Hydrogenophaga atypica sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol 55:341–344
Kato S, Kikuchi S, Kashiwabara T, Takahashi Y, Suzuki K, Itoh T, Ohkuma M, Yamagishi A (2012) Prokaryotic abundance and community composition in a freshwater iron-rich microbial mat at circumneutral pH. Geomicrobiol J 29:896–905
Kellermann C, Griebler C (2009) Thiobacillus thiophilus sp. nov., a chemolithoautotrophic, thiosulfate-oxidizing bacterium isolated from contaminated aquifer sediments. Int J Syst Evol Microbiol 59:583–588
Klatt CG, Wood JM, Rusch DB, Bateson MM, Hamamura N, Heidelberg JF, Grossman AR, Bhaya D, Cohan FM, Kühl M, Bryant DA, Ward DM (2011) Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J 5:1262–1278
Knittel K, Boetius A (2009) Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334
Kodama Y, Watanabe K (2004) Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int J Syst Evol Microbiol 54:2297–2300
Kubo K, Lloyd KG, Biddle J, Amann R, Teske A, Knittel K (2012) Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J 6:1949–1965
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lee SY, Lee MH, Oh TK, Yoon JH (2006) Lutibacter aestuarii sp. nov., isolated from a tidal flat sediment, and emended description of the genus Lutibacter Choi and Cho. Int J Syst Evol Microbiol 62:420–424
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann NY Acad Sci 1125:171–189
Liu J, Wang Z, Belchik SM, Edwards MJ, Liu C, Kennedy DW, Merkley ED, Lipton MD, Butt JN, Richardson DJ, Zachara JM, Fredrickson JK, Rosso KM, Shi L (2012) Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front Microbiol 3:37
Lomans BP, Maas R, Luderer R, Op den Camp HJ, Pol A, van der Drift C, Vogels GD (1999) Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl Environ Microbiol 65:3641–3650
Miller SR, Strong AL, Jones KL, Ungerer MC (2009) Bar-coded pyrosequencing reveals shared bacterial community properties along the temperature gradients of two alkaline hot springs in Yellowstone National Park. Appl Environ Microbiol 75:4565–4572
Mkrtchyan SS (1969) Geology of Armenian SSR, vol IX. Publishing House of AS of ASSR, Yerevan (in Russian)
Neuendorf KKE, Mehl JP Jr, Jackson JA (2005) Glossary of geology, 5th edn. American Geological Institute, Alexandria
Panosyan HH (2010) Phylogenetic diversity based on 16S rRNA gene sequence analysis of aerobic thermophilic endospore-forming bacteria isolated from geothermal springs in Armenia. Biol J Armen 62:73–80
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Robertson CE, Spear JR, Harris JK, Pace NR (2009) Diversity and stratification of archaea in a hypersaline microbial mat. Appl Environ Microbiol 75:1801–1810
Schleper C, Jurgens G, Jonuscheit M (2005) Genomic studies of uncultivated archaea. Nat Rev Microbiol 3:479–488
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Spear JR, Walker JJ, McCollom TM, Pace NR (2005) Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc Natl Acad Sci 102:2555–2560
Stevens TO, McKinley JP (1995) Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270:450–454
Takai K, Horikoshi K (1999) Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285–1297
Takai K, Nakamura K, Toki T, Tsunogai U, Miyazaki M, Miyazaki J, Hirayama H, Nakagawa S, Nunoura T, Horikoshi K (2008) Cell proliferation at 122 degrees C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc Natl Acad Sci USA 5:10949–10954
Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA 108:8420–8425
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Whitman WB, Shieh J, Sohn S, Caras DS, Premachandran U (1986) Isolation and characterization of 22 mesophilic methanococci. Syst Appl Microbiol 7:235–240
Whitman WB, Bowen TL, Boone DR (2006) The methanogenic bacteria. In: Dworkin M (ed) The prokaryotes: archaea and bacteria: firmicutes, actinomyces, vol 3. Springer, New York, pp 165–207
Zeikus JG, Ben-Bassat A, Hegge PW (1980) Microbiology of methanogenesis in thermal, volcanic environments. J Bacteriol 143:432–440
Zhang CL, Pearson A, Li YL, Mills G, Wiegel J (2006) Thermophilic temperature optimum for crenarchaeol synthesis and its implication for archaeal evolution. Appl Environ Microbiol 72:4419–4422
Zhang CL, Ye Q, Huang Z, Li W, Chen J, Song Z, Zhao W, Bagwell C, Inskeep WP, Ross C, Gao L, Wiegel J, Romanek CS, Shock EL, Hedlund BP (2008) Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl Environ Microbiol 74:6417–6426
Acknowledgments
We thank members of the Hedlund lab for advice and discussion. This work was supported by the U.S. National Science Foundation (OISE-0968421), the Armenian National Foundation of Science and Advanced Technologies, and the U.S. Civilian Research & Development Foundation (Grant Number TFP-12-05). B.P.H. acknowledges generous support from Greg Fullmer through the UNLV Foundation.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hedlund, B.P., Dodsworth, J.A., Cole, J.K. et al. An integrated study reveals diverse methanogens, Thaumarchaeota, and yet-uncultivated archaeal lineages in Armenian hot springs. Antonie van Leeuwenhoek 104, 71–82 (2013). https://doi.org/10.1007/s10482-013-9927-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9927-z