Abstract
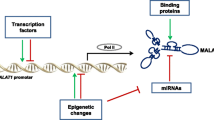
The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a bona fide long noncoding RNA (lncRNA). MALAT1, also known as nuclear-enriched transcript 2 (NEAT2), was discovered as a prognostic marker for lung cancer metastasis but also has been linked to several other human tumor entities. Recent work established a critical regulatory function of this lncRNA in lung cancer metastasis and cell migration. Moreover, MALAT1 is an interesting target for antimetastatic therapy in non-small cell lung carcinoma. Two alternative modes of action have been proposed for MALAT1: regulation of gene expression or alternative splicing. Although the exact mechanism of action in different physiological and pathological conditions still needs to be elucidated, MALAT1 acts as a regulator of gene expression. Although MALAT1 is highly evolutionary conserved in mammals and plays an important role in cancer and metastasis, MALAT1 is not essential for development in a knockout mouse model under normal physiological conditions. Hence, one central question for the future is finding the right stressor and the pathological or environmental condition which requires MALAT1 expression in vivo and entailing its strong evolutionary conservation. Here, we summarize the current knowledge about this important lncRNA. We introduce its discovery, biogenesis, and regulation and describe its known functions, mechanisms of action, and interaction partners.


Similar content being viewed by others
References
Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E et al (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22:8031–8041
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA et al (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938
Wilusz JE, Freier SM, Spector DL (2008) 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135:919–932
Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8:39
Koshimizu TA, Fujiwara Y, Sakai N, Shibata K, Tsuchiya H (2010) Oxytocin stimulates expression of a noncoding RNA tumor marker in a human neuroblastoma cell line. Life Sci 86:455–460
Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL (2009) MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19:347–359
Wilusz JE, Spector DL (2010) An unexpected ending: noncanonical 3′ end processing mechanisms. RNA 16:259–266
Wilusz JE, Jnbaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA (2012) A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26:2392–2407
Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA (2012) Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A 109:19202–19207
Tani H, Nakamura Y, Ijiri K, Akimitsu N (2010) Stability of MALAT-1, a nuclear long non-coding RNA in mammalian cells, varies in various cancer cells. Drug Discov Ther 4:235–239
Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R (2009) Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res 37:e115
Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS (2012) Genome-wide analysis of long noncoding RNA stability. Genome Res 22:885–898
Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F et al (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29:3082–3093
Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C et al (2011) Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 14:459–468
Buratti E, Baralle FE (2008) Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci 13:867–878
Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS (2009) Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res 37:1799–1808
Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V et al (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14:452–458
Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Caceres JF (2012) DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol 19:760–766
Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G (2009) Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136:496–507
Gutschner T, Diederichs S (2012) The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 9:703–719
Schmidt LH, Spieker T, Koschmieder S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L et al (2011) The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 6:1984–1992
Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N (2010) MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett 584:4575–4580
Gutschner T, Baas M, Diederichs S (2011) Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res 21:1944–1954
Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M et al (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73:1180–1189
Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M et al (2012) Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 9:1076–1087
Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV (2012) Malat1 is not an essential component of nuclear speckles in mice. RNA 18:1487–1499
Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C et al (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2:111–123
Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP et al (2006) Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology 44:1012–1024
Lin R, Maeda S, Liu C, Karin M, Edgington TS (2007) A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26:851–858
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS (2012) Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol 29:1810–1816
Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ et al (2009) A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics 10:163
Praz V, Jagannathan V, Bucher P (2004) CleanEx: a database of heterogeneous gene expression data based on a consistent gene nomenclature. Nucleic Acids Res 32:D542–D547
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC et al (2012) Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486:353–360
Guo F, Li Y, Liu Y, Wang J, Li G (2010) Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 42:224–229
Yamada K, Kano J, Tsunoda H, Yoshikawa H, Okubo C, Ishiyama T, Noguchi M (2006) Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci 97:106–112
Xu C, Yang M, Tian J, Wang X, Li Z (2011) MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol 39:169–175
Han Y, Liu Y, Nie L, Gui Y, Cai Z (2013) Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology 81(209):e201–e207
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F (2012) Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst 8:2289–2294
Fellenberg J, Bernd L, Delling G, Witte D, Zahlten-Hinguranage A (2007) Prognostic significance of drug-regulated genes in high-grade osteosarcoma. Mod Pathol 20:1085–1094
Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M et al (2003) Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A 100:6051–6056
Spector DL, Lamond AI (2011) Nuclear speckles. Cold Spring Harb Perspect Biol 3(2):a000646
Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T et al (2012) Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18:738–751
Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726
Chen LL, Carmichael GG (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35:467–478
Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T (2012) Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 31:4020–4034
Nakagawa S, Naganuma T, Shioi G, Hirose T (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193:31–39
Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM (2012) The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13:R17
Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y (2009) Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res 19:381–394
Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147:773–788
Bourgeois CF, Lejeune F, Stevenin J (2004) Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol 78:37–88
Long JC, Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417:15–27
Lin R, Roychowdhury-Saha M, Black C, Watt AT, Marcusson EG, Freier SM, Edgington TS (2011) Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett 585:671–676
Cartegni L, Krainer AR (2003) Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol 10:120–125
Moroy T, Heyd F (2007) The impact of alternative splicing in vivo: mouse models show the way. RNA 13:1155–1171
Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD (2006) Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26:2560–2569
Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG et al (2008) Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A 105:19199–19204
Schuettengruber B, Martinez AM, Iovino N, Cavalli G (2011) Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol 12:799–814
Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF et al (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22:1298–1306
Wagner EJ, Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13:115–126
Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N (2011) Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell 43:340–352
Li L, Feng T, Lian Y, Zhang G, Garen A, Song X (2009) Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci U S A 106:12956–12961
Grippo PJ, Sandgren EP (2000) Highly invasive transitional cell carcinoma of the bladder in a simian virus 40 T-antigen transgenic mouse model. Am J Pathol 157:805–813
Acknowledgments
We apologize to all scientists whose important work could not be cited in this review due to space constraints. Our research is supported by the German Research Foundation (DFG Transregio TRR77, TP B03), the Marie Curie Program of the European Commission, the Helmholtz Society (VH-NG-504), the Virtual Helmholtz Institute for Resistance in Leukemia, the German Cancer Research Center (DKFZ), and the Institute of Pathology, University of Heidelberg.
Conflict of interest
The authors declare no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Parts of this text have been included in the doctoral thesis of Tony Gutschner.
Rights and permissions
About this article
Cite this article
Gutschner, T., Hämmerle, M. & Diederichs, S. MALAT1 — a paradigm for long noncoding RNA function in cancer. J Mol Med 91, 791–801 (2013). https://doi.org/10.1007/s00109-013-1028-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-013-1028-y